Análise Multivaridada das Variáveis Químicas do Solo
Walmes M. Zeviani & Milson E. Serafim
Source:vignettes/teca_qui.Rmd
teca_qui.RmdAnálise Exploratória
#-----------------------------------------------------------------------
# Carrega os pacotes necessários.
library(lattice)
library(latticeExtra)
library(plyr)
library(reshape2)
library(EACS)#-----------------------------------------------------------------------
# Análise exploratório dos dados.
# Pra facilitar o manuseio, vamos usar um nome curto.
qui <- teca_qui
str(qui)## 'data.frame': 150 obs. of 15 variables:
## $ loc: int 1 1 1 2 2 2 3 3 3 4 ...
## $ cam: Factor w/ 3 levels "[0, 5)","[5, 40)",..: 1 2 3 1 2 3 1 2 3 1 ...
## $ ph : num 6.8 6.7 6.7 4.7 4.7 4.9 7.6 6.8 6.9 6.6 ...
## $ p : num 22.51 0.83 0.01 3.89 0.69 ...
## $ k : num 72.24 13.42 7.23 48.13 12.34 ...
## $ ca : num 8.27 2.91 2.33 0.97 0.76 0.21 7.63 3.22 2.22 5.54 ...
## $ mg : num 1.7 1.77 0.51 0.16 0.14 0 1.53 1.31 1.09 1.77 ...
## $ al : num 0 0 0 0.3 0.6 0.6 0 0 0 0 ...
## $ ctc: num 12.47 6.57 4.52 5.3 4.17 ...
## $ sat: num 81.4 71.7 63.2 23.7 22.4 ...
## $ mo : num 72.2 25.6 9.7 34.4 8.7 9.7 50.4 15.6 9.5 50.2 ...
## $ arg: num 184 215 286 232 213 ...
## $ are: num 770 750 674 741 775 ...
## $ cas: num 1.8 2.2 3.7 0.4 1.1 1.7 8.4 19 14.3 5.5 ...
## $ acc: num 770 750 676 741 775 ...#-----------------------------------------------------------------------
# splom(qui[, -(1:2)])
# Pares de diagramas de dispersão para as variáveis químicas.
splom(qui[, 3:11])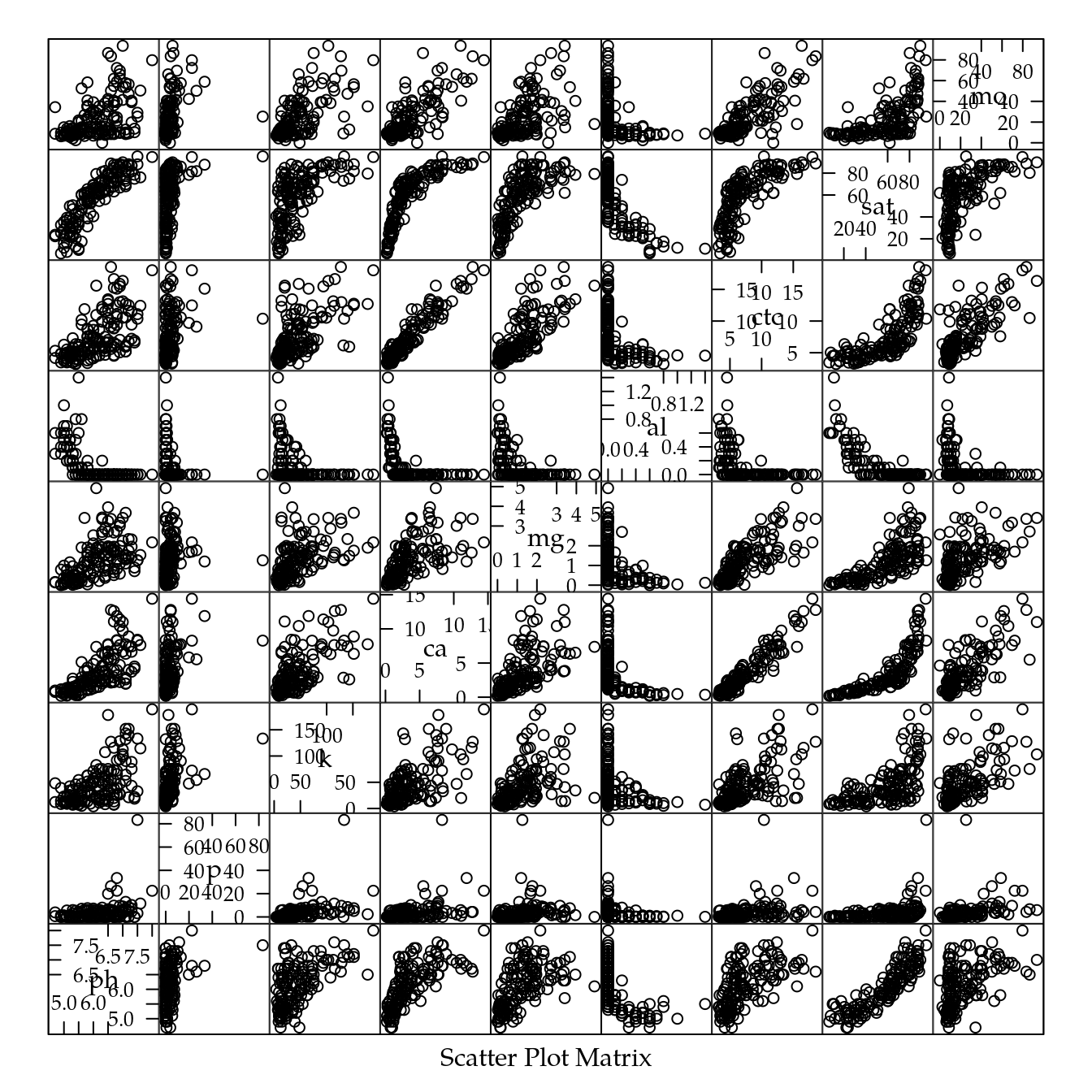
# Pares de diagramas de dispersão para as variáveis físicas.
splom(qui[, 11:15])
#-----------------------------------------------------------------------
# Agregando para a média das 3 camadas em cada local.
quim <- ddply(qui[, -2], .(loc), .fun = colMeans)
str(quim)## 'data.frame': 50 obs. of 14 variables:
## $ loc: num 1 2 3 4 5 6 7 8 9 10 ...
## $ ph : num 6.73 4.77 7.1 5.93 5.63 ...
## $ p : num 7.78 1.56 4.53 9.27 1.43 ...
## $ k : num 31 23 55.7 26.3 30.9 ...
## $ ca : num 4.503 0.647 4.357 2.847 1.647 ...
## $ mg : num 1.327 0.1 1.31 0.713 0.893 ...
## $ al : num 0 0.5 0 0.133 0.133 ...
## $ ctc: num 7.85 4.31 7.91 5.95 5.01 ...
## $ sat: num 72.1 17.6 72.1 53 45.8 ...
## $ mo : num 35.8 17.6 25.2 22.8 19.9 ...
## $ arg: num 228 227 218 190 330 ...
## $ are: num 731 756 696 750 637 ...
## $ cas: num 2.57 1.07 13.9 6.6 7.4 ...
## $ acc: num 732 756 700 752 640 ...splom(quim[, 2:10])
splom(quim[, 10:14])
Análise de Componentes Principais
Camadas como indivíduos
#-----------------------------------------------------------------------
# Cria uma matriz só com as variáveis de solo.
quiv <- qui[, -(1:2)]
rownames(quiv) <- with(qui, paste(loc, as.roman(as.numeric(cam)),
sep = "-"))
acp <- princomp(quiv, cor = TRUE)
summary(acp)## Importance of components:
## Comp.1 Comp.2 Comp.3 Comp.4
## Standard deviation 2.3854986 1.7376345 1.07551158 0.93098921
## Proportion of Variance 0.4377388 0.2322595 0.08897886 0.06667238
## Cumulative Proportion 0.4377388 0.6699983 0.75897712 0.82564950
## Comp.5 Comp.6 Comp.7 Comp.8
## Standard deviation 0.88988785 0.66322487 0.62727651 0.56409125
## Proportion of Variance 0.06091541 0.03383594 0.03026737 0.02447684
## Cumulative Proportion 0.88656491 0.92040085 0.95066823 0.97514507
## Comp.9 Comp.10 Comp.11
## Standard deviation 0.43024624 0.263392164 0.222279957
## Proportion of Variance 0.01423937 0.005336572 0.003800645
## Cumulative Proportion 0.98938444 0.994721010 0.998521655
## Comp.12 Comp.13
## Standard deviation 0.1056119374 0.0898031581
## Proportion of Variance 0.0008579909 0.0006203544
## Cumulative Proportion 0.9993796456 1.0000000000# Proporção de variância acumulada.
plot(cbind(x = 0:length(acp$sdev),
y = c(0, cumsum(acp$sdev^2))/sum(acp$sdev^2)),
type = "o",
xlab = "Componente",
ylab = "Proporção de variância acumulada")
abline(h = 0.75, lty = 2)
#-----------------------------------------------------------------------
# Carregamentos.
# acp$loadings
# A fração dos carregamentos mais importantes.
imp <- function(x, f = 0.25) {
a <- abs(x)
k <- ceiling(f * length(x))
i <- sort(a, decreasing = TRUE)[k]
x[a <= i] <- NA
return(x)
}
apply(acp$loadings[, 1:4], MARGIN = 2, FUN = imp, f = 0.25)## Comp.1 Comp.2 Comp.3 Comp.4
## ph NA NA NA NA
## p NA NA NA 0.3540011
## k NA NA NA NA
## ca 0.3850495 NA NA NA
## mg NA NA NA NA
## al NA NA 0.1493058 -0.5714961
## ctc 0.3772880 NA NA -0.3878400
## sat 0.3906511 NA NA NA
## mo NA NA NA NA
## arg NA 0.5518568 NA NA
## are NA -0.5488643 NA NA
## cas NA NA -0.8918405 NA
## acc NA -0.5048946 -0.3669276 NA#-----------------------------------------------------------------------
# Gráficos biplot.
biplot(acp, choices = c(1, 2))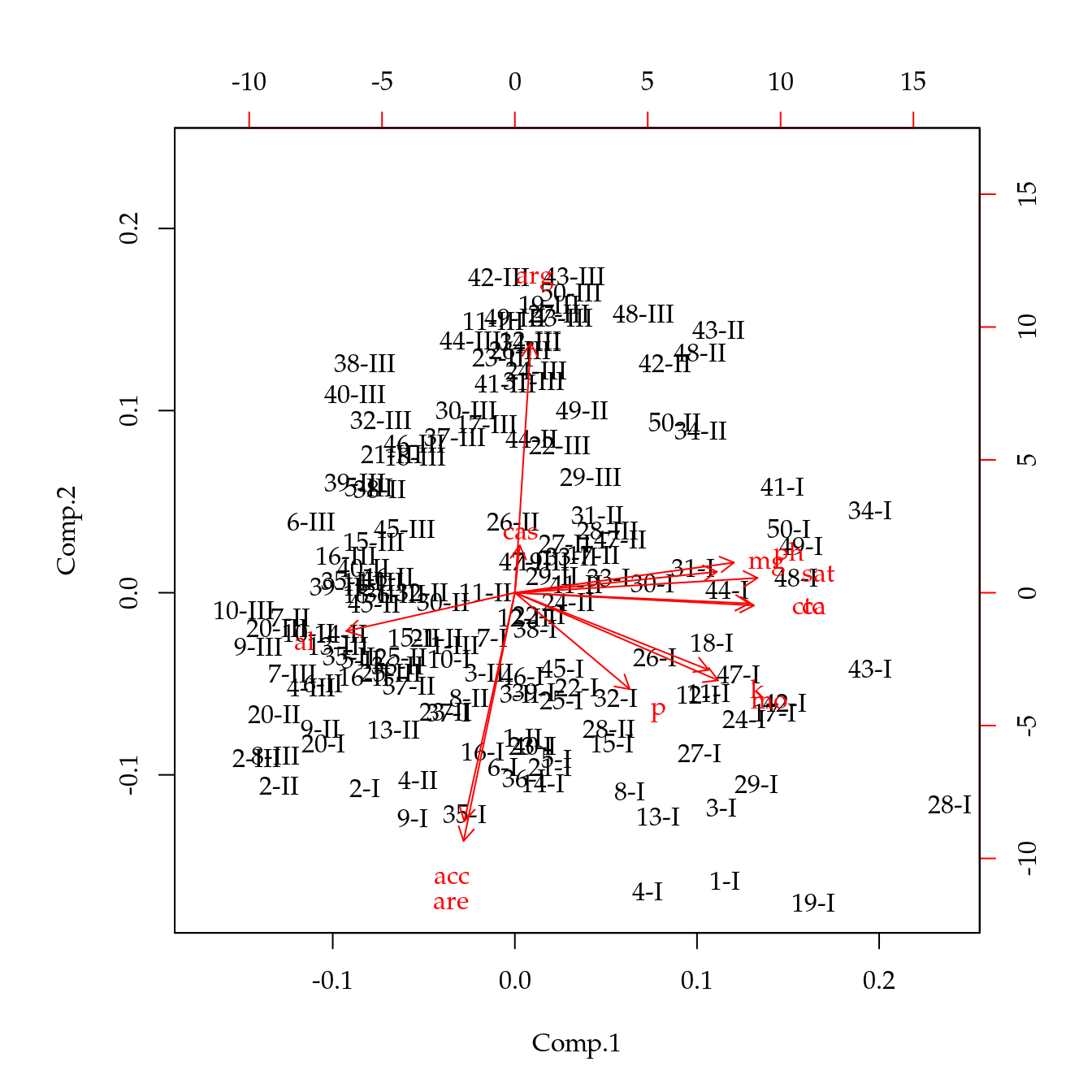

#-----------------------------------------------------------------------
# Indentifição das camadas.
# Escores.
sc <- acp$scores
pair <- c(1, 2)
# Aplitudes dos escores.
rgs <- apply(sc[, pair], MARGIN = 2, FUN = range)
# Carregamentos.
ld <- acp$loadings
# Amplitude dos carregamentos.
rgl <- apply(ld[, pair], MARGIN = 2, FUN = range)
# Fator de encolhimento para colocar setas em meio aos pontos.
f <- 0.5
xsc <- f * max(abs(rgs[, 1]))/max(abs(rgl[, 1]))
ysc <- f * max(abs(rgs[, 2]))/max(abs(rgl[, 2]))
par(pty = "s")
plot(rgs, col = "white", asp = 1,
xlab = names(rgs)[1], ylab = names(rgs)[2])
points(sc[, pair], asp = 1, col = as.numeric(qui$cam), pch = 19)
abline(v = 0, h = 0, lty = 2)
arrows(x0 = rep(0, 1),
y0 = rep(0, 1),
x1 = ld[, pair[1]] * xsc,
y1 = ld[, pair[2]] * ysc,
length = 0.1,
col = 2)
text(x = ld[, pair[1]] * xsc * 1.1,
y = ld[, pair[2]] * ysc * 1.1,
labels = rownames(ld))
legend("topleft", legend = levels(qui$cam),
pch = 19, col = 1:3, bty = "n")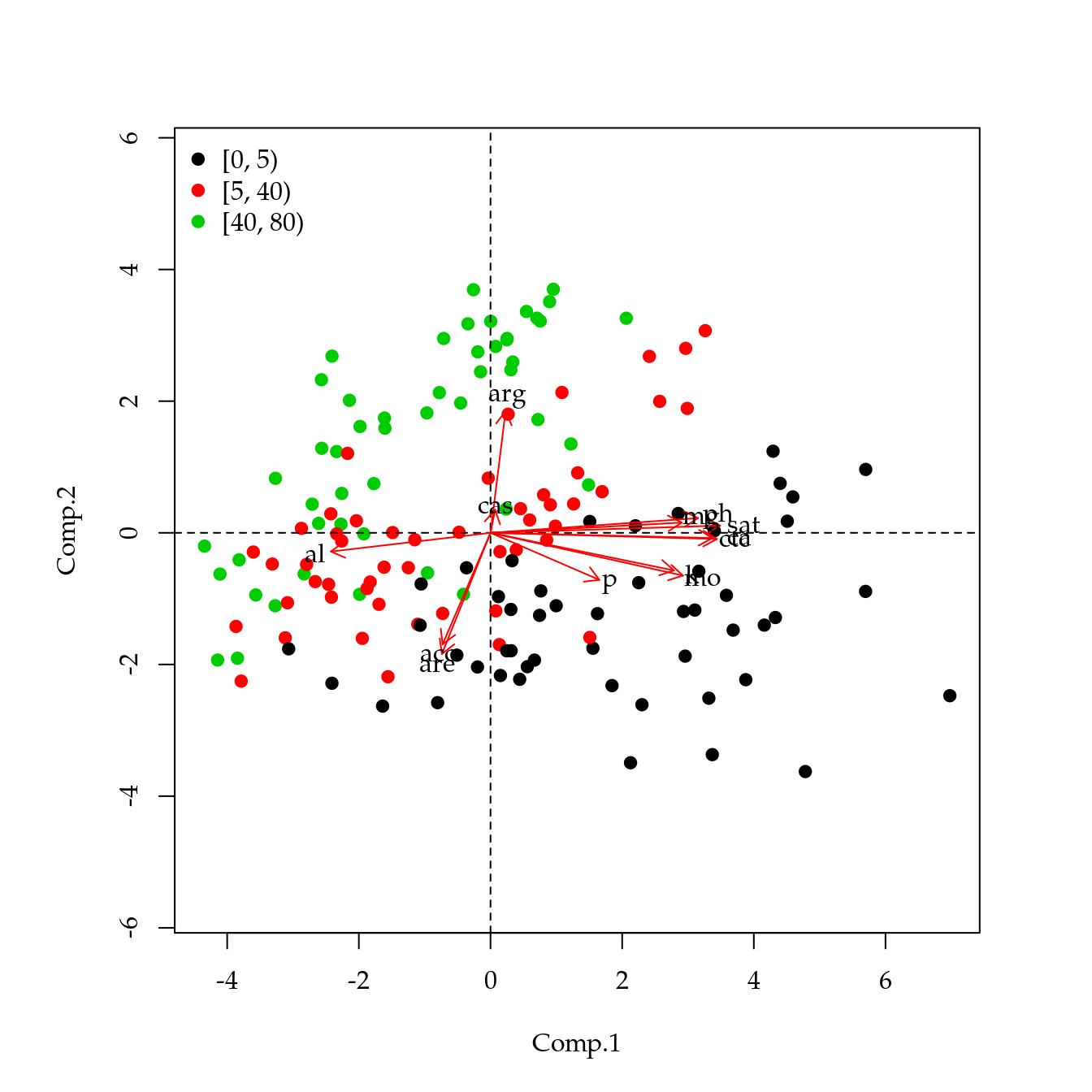
par(pty = "m")Camadas como variáveis
#-----------------------------------------------------------------------
# Empilhar as variáveis de solo.
quie <- melt(data = qui,
id.vars = c("loc", "cam"))
str(quie)## 'data.frame': 1950 obs. of 4 variables:
## $ loc : int 1 1 1 2 2 2 3 3 3 4 ...
## $ cam : Factor w/ 3 levels "[0, 5)","[5, 40)",..: 1 2 3 1 2 3 1 2 3 1 ...
## $ variable: Factor w/ 13 levels "ph","p","k","ca",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ value : num 6.8 6.7 6.7 4.7 4.7 4.9 7.6 6.8 6.9 6.6 ...quie$camvar <- with(quie, paste(as.character(variable),
as.character(as.roman(as.integer(cam))),
sep = ":"))
# Desempilhar as variáveis.
quie <- dcast(data = quie, formula = loc ~ camvar, value.var = "value")
str(quie)## 'data.frame': 50 obs. of 40 variables:
## $ loc : int 1 2 3 4 5 6 7 8 9 10 ...
## $ acc:I : num 770 741 718 791 734 ...
## $ acc:II : num 750 775 704 788 667 ...
## $ acc:III: num 676 752 679 678 518 ...
## $ al:I : num 0 0.3 0 0 0 0 0 0 0 0.1 ...
## $ al:II : num 0 0.6 0 0 0.2 0.5 0.6 0 0.3 0.6 ...
## $ al:III : num 0 0.6 0 0.4 0.2 0.6 0.8 0.7 1 1.4 ...
## $ are:I : num 770 741 716 790 733 ...
## $ are:II : num 750 775 698 788 665 ...
## $ are:III: num 674 752 674 674 514 ...
## $ arg:I : num 184 232 193 142 236 ...
## $ arg:II : num 215 213 234 169 304 ...
## $ arg:III: num 286 237 226 258 452 ...
## $ ca:I : num 8.27 0.97 7.63 5.54 2.99 1.88 1.6 5.82 1.91 1.73 ...
## $ ca:II : num 2.91 0.76 3.22 2.25 0.93 0.7 0.24 2.39 1.15 0.72 ...
## $ ca:III : num 2.33 0.21 2.22 0.75 1.02 0.3 0.6 0.28 0.47 0.36 ...
## $ cas:I : num 1.8 0.4 8.4 5.5 5.5 20.9 12.4 2 1.4 0.8 ...
## $ cas:II : num 2.2 1.1 19 3.1 7.2 23.4 2.9 2.1 2.5 1.6 ...
## $ cas:III: num 3.7 1.7 14.3 11.2 9.5 31.8 5 3.6 3.3 1.4 ...
## $ ctc:I : num 12.47 5.3 12.41 9.11 7.57 ...
## $ ctc:II : num 6.57 4.17 6.26 4.56 4.1 5.16 5.02 5.28 4.86 3.86 ...
## $ ctc:III: num 4.52 3.47 5.06 4.18 3.37 4.48 4.53 3.61 4.58 4.54 ...
## $ k:I : num 72.2 48.1 114.5 54.3 56.8 ...
## $ k:II : num 13.4 12.3 25.8 11.1 22.2 ...
## $ k:III : num 7.23 8.64 26.83 13.57 13.57 ...
## $ mg:I : num 1.7 0.16 1.53 1.77 2.12 1.53 1.42 1.67 0.21 0.69 ...
## $ mg:II : num 1.77 0.14 1.31 0.21 0.52 0.36 0.14 0.99 0.05 0.37 ...
## $ mg:III : num 0.51 0 1.09 0.16 0.04 0.1 0.18 0.28 0.04 0.12 ...
## $ mo:I : num 72.2 34.4 50.4 50.2 39.1 35 28.6 42.3 19.9 24.3 ...
## $ mo:II : num 25.6 8.7 15.6 10 11.2 5.8 8.5 11.8 6.3 6.9 ...
## $ mo:III : num 9.7 9.7 9.5 8.2 9.3 9.3 8.2 7.6 7.1 8.8 ...
## $ p:I : num 22.51 3.89 11.52 26.43 3.79 ...
## $ p:II : num 0.83 0.69 1.66 1.17 0.49 0.69 0.21 1.14 1.36 1.04 ...
## $ p:III : num 0.01 0.1 0.41 0.2 0.01 0.1 0.01 0.01 1.07 0.1 ...
## $ ph:I : num 6.8 4.7 7.6 6.6 6 5.8 5.9 6.5 5.5 5.8 ...
## $ ph:II : num 6.7 4.7 6.8 6.2 5.1 5.2 5.1 6.4 4.9 5 ...
## $ ph:III : num 6.7 4.9 6.9 5 5.8 5.5 5.6 5.1 5 5.5 ...
## $ sat:I : num 81.4 23.7 76.1 81.8 69.4 ...
## $ sat:II : num 71.7 22.4 73.4 54.5 36.8 ...
## $ sat:III: num 63.23 6.69 66.78 22.59 31.26 ...#-----------------------------------------------------------------------
acp <- princomp(quie, cor = TRUE)
# summary(acp)
# Proporção de variância acumulada.
plot(cbind(x = 0:length(acp$sdev),
y = c(0, cumsum(acp$sdev^2))/sum(acp$sdev^2)),
type = "o",
xlab = "Componente",
ylab = "Proporção de variância acumulada")
abline(h = 0.75, lty = 2)
#-----------------------------------------------------------------------
# Carregamentos.
# acp$loadings
apply(acp$loadings[, 1:4], MARGIN = 2, FUN = imp, f = 0.25)## Comp.1 Comp.2 Comp.3 Comp.4
## loc NA NA 0.1908126 NA
## acc:I NA -0.2238693 NA NA
## acc:II NA NA 0.2270238 -0.1622790
## acc:III NA NA NA -0.4100126
## al:I NA NA NA NA
## al:II NA NA NA NA
## al:III NA NA NA NA
## are:I NA NA NA NA
## are:II NA -0.2585851 NA NA
## are:III NA NA NA -0.2484359
## arg:I NA 0.2307062 NA NA
## arg:II NA 0.2516177 NA NA
## arg:III NA NA NA 0.3462011
## ca:I 0.2120379 NA NA NA
## ca:II 0.2024972 NA NA NA
## ca:III 0.2020180 NA NA NA
## cas:I NA NA 0.4528881 NA
## cas:II NA NA 0.4912041 NA
## cas:III NA NA 0.4324992 -0.1790642
## ctc:I 0.2128865 NA NA NA
## ctc:II 0.2094659 NA NA NA
## ctc:III NA NA 0.1545793 NA
## k:I NA NA NA NA
## k:II NA -0.2500313 0.1710886 NA
## k:III NA -0.2891170 0.1546419 NA
## mg:I NA NA -0.1973335 NA
## mg:II 0.1856483 NA NA NA
## mg:III NA NA NA NA
## mo:I NA NA NA -0.1657718
## mo:II NA NA NA NA
## mo:III NA NA NA NA
## p:I NA NA NA NA
## p:II NA -0.2969129 NA 0.3373342
## p:III NA -0.2756235 NA 0.3858953
## ph:I NA -0.2149295 NA NA
## ph:II NA NA NA -0.1686722
## ph:III NA NA NA NA
## sat:I 0.1924693 NA NA NA
## sat:II 0.2049075 NA NA NA
## sat:III 0.2144155 NA NA NA#-----------------------------------------------------------------------
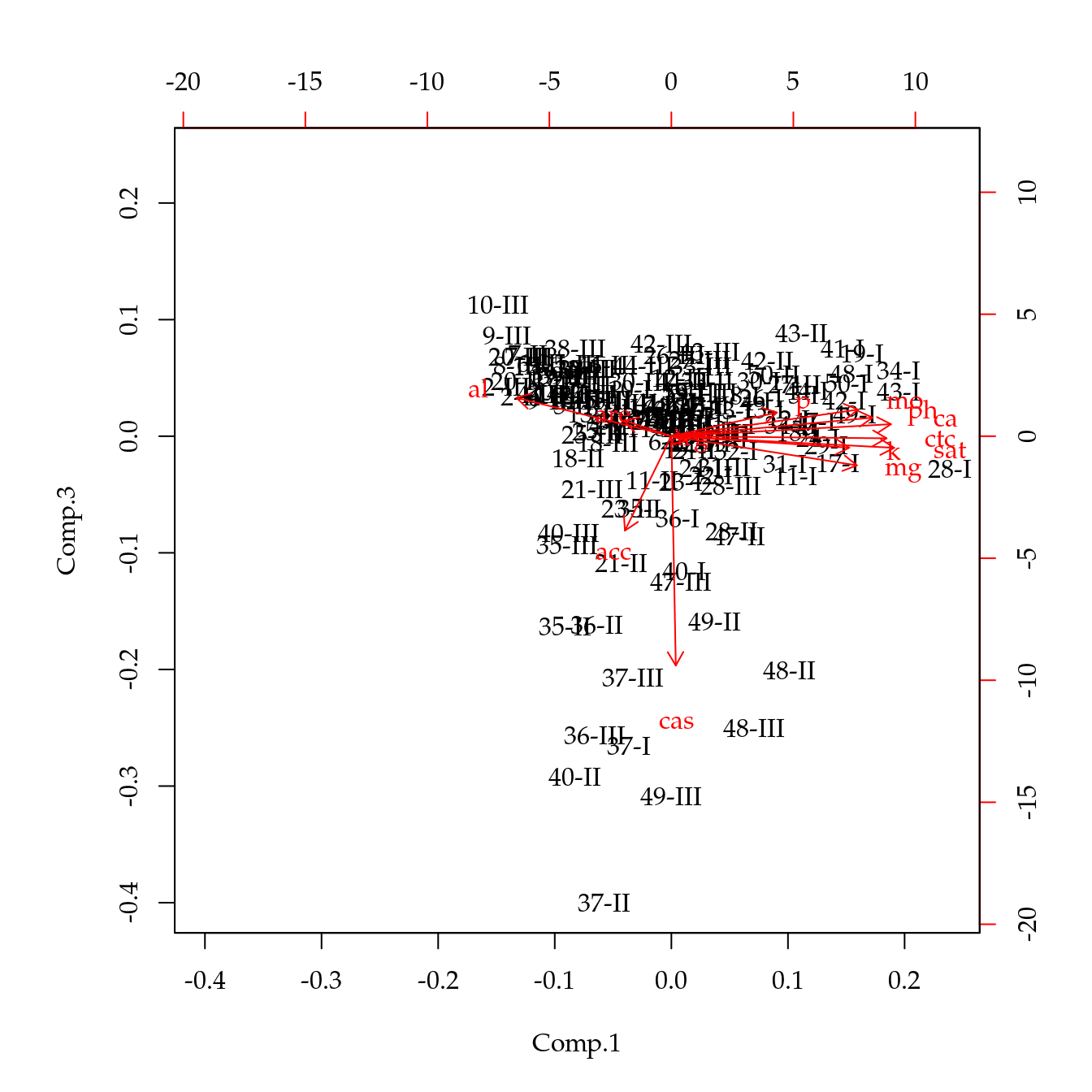
# Gráficos biplot.
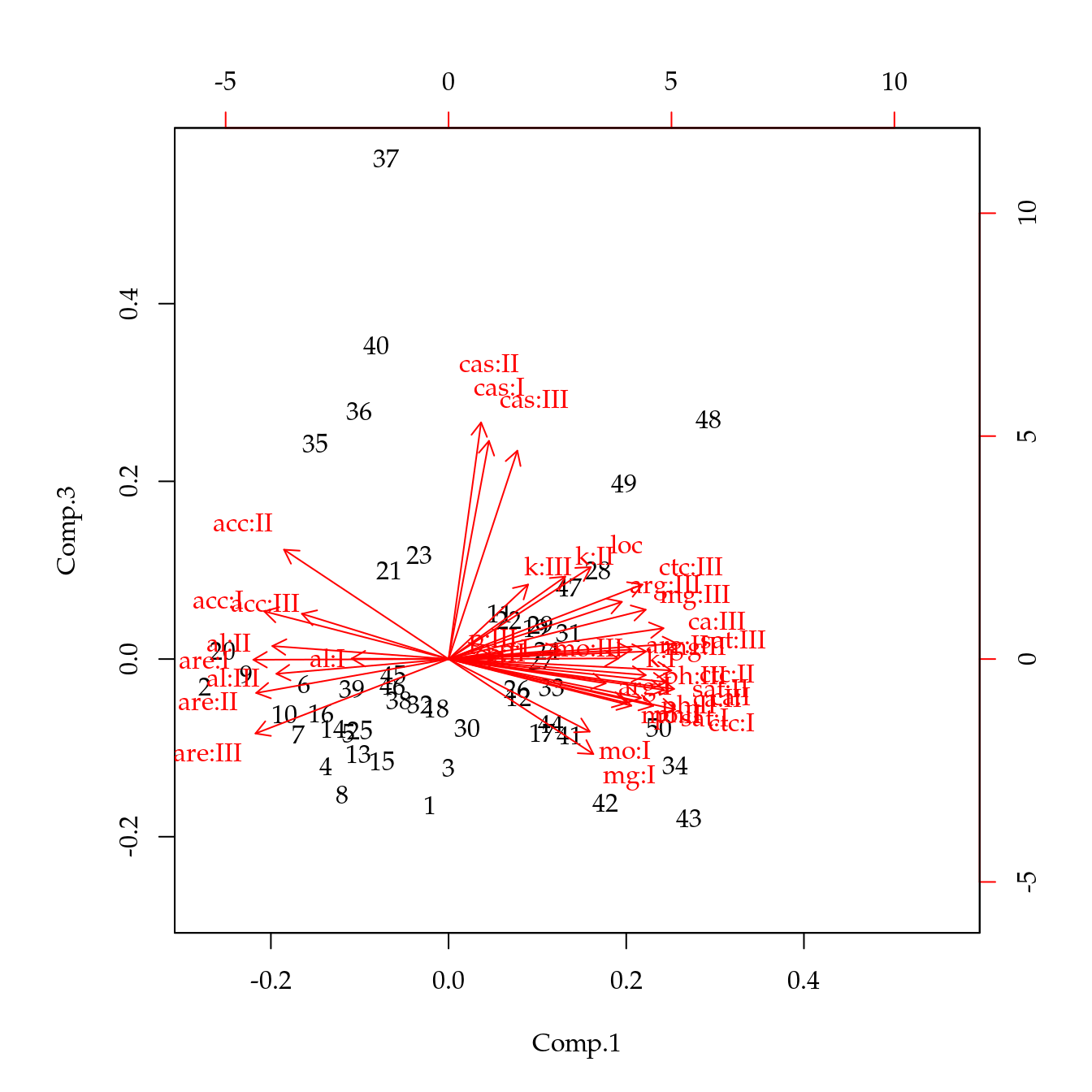
biplot(acp, choices = c(1, 2))

Informações da Sessão
## Atualizado em 11 de julho de 2019.
##
## R version 3.6.1 (2019-07-05)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 18.04.2 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.7.1
## LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.7.1
##
## locale:
## [1] LC_CTYPE=pt_BR.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=pt_BR.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=pt_BR.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=pt_BR.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=pt_BR.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods
## [7] base
##
## other attached packages:
## [1] reshape2_1.4.3 plyr_1.8.4 EACS_0.0-7
## [4] wzRfun_0.91 captioner_2.2.3 latticeExtra_0.6-28
## [7] RColorBrewer_1.1-2 lattice_0.20-38 knitr_1.23
##
## loaded via a namespace (and not attached):
## [1] Rcpp_1.0.1 compiler_3.6.1 prettyunits_1.0.2
## [4] remotes_2.1.0 tools_3.6.1 testthat_2.1.1
## [7] digest_0.6.20 pkgbuild_1.0.3 pkgload_1.0.2
## [10] evaluate_0.14 memoise_1.1.0 rlang_0.4.0
## [13] rstudioapi_0.10 cli_1.1.0 commonmark_1.7
## [16] yaml_2.2.0 pkgdown_1.3.0 xfun_0.8
## [19] withr_2.1.2 stringr_1.4.0 roxygen2_6.1.1
## [22] xml2_1.2.0 desc_1.2.0 fs_1.3.1
## [25] devtools_2.1.0 rprojroot_1.3-2 grid_3.6.1
## [28] glue_1.3.1 R6_2.4.0 processx_3.4.0
## [31] rmarkdown_1.13 sessioninfo_1.1.1 callr_3.3.0
## [34] magrittr_1.5 usethis_1.5.1 backports_1.1.4
## [37] ps_1.3.0 htmltools_0.3.6 MASS_7.3-51.4
## [40] assertthat_0.2.1 stringi_1.4.3 crayon_1.3.4